DeepHPMs(Deep Hidden Physics Models)¶
# 案例 1
# linux
wget -nc https://paddle-org.bj.bcebos.com/paddlescience/datasets/DeepHPMs/burgers_sine.mat -P ./datasets/
# windows
# curl https://paddle-org.bj.bcebos.com/paddlescience/datasets/DeepHPMs/burgers_sine.mat --output ./datasets/burgers_sine.mat
python burgers.py DATASET_PATH=./datasets/burgers_sine.mat DATASET_PATH_SOL=./datasets/burgers_sine.mat
# 案例 2
# linux
wget -nc https://paddle-org.bj.bcebos.com/paddlescience/datasets/DeepHPMs/burgers_sine.mat -P ./datasets/
wget -nc https://paddle-org.bj.bcebos.com/paddlescience/datasets/DeepHPMs/burgers.mat -P ./datasets/
# windows
# curl https://paddle-org.bj.bcebos.com/paddlescience/datasets/DeepHPMs/burgers_sine.mat --output ./datasets/burgers_sine.mat
# curl https://paddle-org.bj.bcebos.com/paddlescience/datasets/DeepHPMs/burgers.mat --output ./datasets/burgers.mat
python burgers.py DATASET_PATH=./datasets/burgers_sine.mat DATASET_PATH_SOL=./datasets/burgers.mat
# 案例 3
# linux
wget -nc https://paddle-org.bj.bcebos.com/paddlescience/datasets/DeepHPMs/burgers.mat -P ./datasets/
wget -nc https://paddle-org.bj.bcebos.com/paddlescience/datasets/DeepHPMs/burgers_sine.mat -P ./datasets/
# windows
# curl https://paddle-org.bj.bcebos.com/paddlescience/datasets/DeepHPMs/burgers.mat --output ./datasets/burgers.mat
# curl https://paddle-org.bj.bcebos.com/paddlescience/datasets/DeepHPMs/burgers_sine.mat --output ./datasets/burgers_sine.mat
python burgers.py DATASET_PATH=./datasets/burgers.mat DATASET_PATH_SOL=./datasets/burgers_sine.mat
# 案例 4
# linux
wget -nc https://paddle-org.bj.bcebos.com/paddlescience/datasets/DeepHPMs/KdV_sine.mat -P ./datasets/
# windows
# curl https://paddle-org.bj.bcebos.com/paddlescience/datasets/DeepHPMs/KdV_sine.mat --output ./datasets/KdV_sine.mat
python korteweg_de_vries.py DATASET_PATH=./datasets/KdV_sine.mat DATASET_PATH_SOL=./datasets/KdV_sine.mat
# 案例 5
# linux
wget -nc https://paddle-org.bj.bcebos.com/paddlescience/datasets/DeepHPMs/KdV_sine.mat -P ./datasets/
wget -nc https://paddle-org.bj.bcebos.com/paddlescience/datasets/DeepHPMs/KdV_cos.mat -P ./datasets/
# windows
# curl https://paddle-org.bj.bcebos.com/paddlescience/datasets/DeepHPMs/KdV_sine.mat --output ./datasets/KdV_sine.mat
# curl https://paddle-org.bj.bcebos.com/paddlescience/datasets/DeepHPMs/KdV_cos.mat --output ./datasets/KdV_cos.mat
python korteweg_de_vries.py DATASET_PATH=./datasets/KdV_sine.mat DATASET_PATH_SOL=./datasets/KdV_cos.mat
# 案例 6
# linux
wget -nc https://paddle-org.bj.bcebos.com/paddlescience/datasets/DeepHPMs/KS.mat -P ./datasets/
# windows
# curl https://paddle-org.bj.bcebos.com/paddlescience/datasets/DeepHPMs/KS.mat --output ./datasets/KS.mat
python kuramoto_sivashinsky.py DATASET_PATH=./datasets/KS.mat DATASET_PATH_SOL=./datasets/KS.mat
# 案例 7
# linux
wget -nc https://paddle-org.bj.bcebos.com/paddlescience/datasets/DeepHPMs/cylinder.mat -P ./datasets/
# windows
# curl https://paddle-org.bj.bcebos.com/paddlescience/datasets/DeepHPMs/cylinder.mat --output ./datasets/cylinder.mat
python navier_stokes.py DATASET_PATH=./datasets/cylinder.mat DATASET_PATH_SOL=./datasets/cylinder.mat
# 案例 8
# linux
wget -nc https://paddle-org.bj.bcebos.com/paddlescience/datasets/DeepHPMs/NLS.mat -P ./datasets/
# windows
# curl https://paddle-org.bj.bcebos.com/paddlescience/datasets/DeepHPMs/NLS.mat --output ./datasets/NLS.mat
python schrodinger.py DATASET_PATH=./datasets/NLS.mat DATASET_PATH_SOL=./datasets/NLS.mat
# 案例 1
# linux
wget -nc https://paddle-org.bj.bcebos.com/paddlescience/datasets/DeepHPMs/burgers_sine.mat -P ./datasets/
# windows
# curl https://paddle-org.bj.bcebos.com/paddlescience/datasets/DeepHPMs/burgers_sine.mat --output ./datasets/burgers_sine.mat
python burgers.py mode=eval DATASET_PATH=./datasets/burgers_sine.mat DATASET_PATH_SOL=./datasets/burgers_sine.mat EVAL.pretrained_model_path=https://paddle-org.bj.bcebos.com/paddlescience/models/DeepHPMs/burgers_same_pretrained.pdparams
# 案例 2
# linux
wget -nc https://paddle-org.bj.bcebos.com/paddlescience/datasets/DeepHPMs/burgers_sine.mat -P ./datasets/
wget -nc https://paddle-org.bj.bcebos.com/paddlescience/datasets/DeepHPMs/burgers.mat -P ./datasets/
# windows
# curl https://paddle-org.bj.bcebos.com/paddlescience/datasets/DeepHPMs/burgers_sine.mat --output ./datasets/burgers_sine.mat
# curl https://paddle-org.bj.bcebos.com/paddlescience/datasets/DeepHPMs/burgers.mat --output ./datasets/burgers.mat
python burgers.py mode=eval DATASET_PATH=./datasets/burgers_sine.mat DATASET_PATH_SOL=./datasets/burgers.mat EVAL.pretrained_model_path=https://paddle-org.bj.bcebos.com/paddlescience/models/DeepHPMs/burgers_diff_pretrained.pdparams
# 案例 3
# linux
wget -nc https://paddle-org.bj.bcebos.com/paddlescience/datasets/DeepHPMs/burgers.mat -P ./datasets/
wget -nc https://paddle-org.bj.bcebos.com/paddlescience/datasets/DeepHPMs/burgers_sine.mat -P ./datasets/
# windows
# curl https://paddle-org.bj.bcebos.com/paddlescience/datasets/DeepHPMs/burgers.mat --output ./datasets/burgers.mat
# curl https://paddle-org.bj.bcebos.com/paddlescience/datasets/DeepHPMs/burgers_sine.mat --output ./datasets/burgers_sine.mat
python burgers.py mode=eval DATASET_PATH=./datasets/burgers.mat DATASET_PATH_SOL=./datasets/burgers_sine.mat EVAL.pretrained_model_path=https://paddle-org.bj.bcebos.com/paddlescience/models/DeepHPMs/burgers_diff_swap_pretrained.pdparams
# 案例 4
# linux
wget -nc https://paddle-org.bj.bcebos.com/paddlescience/datasets/DeepHPMs/KdV_sine.mat -P ./datasets/
# windows
# curl https://paddle-org.bj.bcebos.com/paddlescience/datasets/DeepHPMs/KdV_sine.mat --output ./datasets/KdV_sine.mat
python korteweg_de_vries.py mode=eval DATASET_PATH=./datasets/KdV_sine.mat DATASET_PATH_SOL=./datasets/KdV_sine.mat EVAL.pretrained_model_path=https://paddle-org.bj.bcebos.com/paddlescience/models/DeepHPMs/kdv_same_pretrained.pdparams
# 案例 5
# linux
wget -nc https://paddle-org.bj.bcebos.com/paddlescience/datasets/DeepHPMs/KdV_sine.mat -P ./datasets/
wget -nc https://paddle-org.bj.bcebos.com/paddlescience/datasets/DeepHPMs/KdV_cos.mat -P ./datasets/
# windows
# curl https://paddle-org.bj.bcebos.com/paddlescience/datasets/DeepHPMs/KdV_sine.mat --output ./datasets/KdV_sine.mat
# curl https://paddle-org.bj.bcebos.com/paddlescience/datasets/DeepHPMs/KdV_cos.mat --output ./datasets/KdV_cos.mat
python korteweg_de_vries.py mode=eval DATASET_PATH=./datasets/KdV_sine.mat DATASET_PATH_SOL=./datasets/KdV_cos.mat EVAL.pretrained_model_path=https://paddle-org.bj.bcebos.com/paddlescience/models/DeepHPMs/kdv_diff_pretrained.pdparams
# 案例 6
# linux
wget -nc https://paddle-org.bj.bcebos.com/paddlescience/datasets/DeepHPMs/KS.mat -P ./datasets/
# windows
# curl https://paddle-org.bj.bcebos.com/paddlescience/datasets/DeepHPMs/KS.mat --output ./datasets/KS.mat
python kuramoto_sivashinsky.py mode=eval DATASET_PATH=./datasets/KS.mat DATASET_PATH_SOL=./datasets/KS.mat EVAL.pretrained_model_path=https://paddle-org.bj.bcebos.com/paddlescience/models/DeepHPMs/ks_pretrained.pdparams
# 案例 7
# linux
wget -nc https://paddle-org.bj.bcebos.com/paddlescience/datasets/DeepHPMs/cylinder.mat -P ./datasets/
# windows
# curl https://paddle-org.bj.bcebos.com/paddlescience/datasets/DeepHPMs/cylinder.mat --output ./datasets/cylinder.mat
python navier_stokes.py mode=eval DATASET_PATH=./datasets/cylinder.mat DATASET_PATH_SOL=./datasets/cylinder.mat EVAL.pretrained_model_path=https://paddle-org.bj.bcebos.com/paddlescience/models/DeepHPMs/ns_pretrained.pdparams
# 案例 8
# linux
wget -nc https://paddle-org.bj.bcebos.com/paddlescience/datasets/DeepHPMs/NLS.mat -P ./datasets/
# windows
# curl https://paddle-org.bj.bcebos.com/paddlescience/datasets/DeepHPMs/NLS.mat --output ./datasets/NLS.mat
python schrodinger.py mode=eval DATASET_PATH=./datasets/NLS.mat DATASET_PATH_SOL=./datasets/NLS.mat EVAL.pretrained_model_path=https://paddle-org.bj.bcebos.com/paddlescience/models/DeepHPMs/schrodinger_pretrained.pdparams
| 序号 | 案例名称 | stage1、2 数据集 | stage3(eval)数据集 | 预训练模型 | 指标 |
|---|---|---|---|---|---|
| 1 | burgers | burgers_sine.mat | burgers_sine.mat | burgers_same_pretrained.pdparams | l2 error: 0.0088 |
| 2 | burgers | burgers_sine.mat | burgers.mat | burgers_diff_pretrained.pdparams | l2 error: 0.0379 |
| 3 | burgers | burgers.mat | burgers_sine.mat | burgers_diff_swap_pretrained.pdparams | l2 error: 0.2904 |
| 4 | korteweg_de_vries | KdV_sine.mat | KdV_sine.mat | kdv_same_pretrained.pdparams | l2 error: 0.0567 |
| 5 | korteweg_de_vries | KdV_sine.mat | KdV_cos.mat | kdv_diff_pretrained.pdparams | l2 error: 0.1142 |
| 6 | kuramoto_sivashinsky | KS.mat | KS.mat | ks_pretrained.pdparams | l2 error: 0.1166 |
| 7 | navier_stokes | cylinder.mat | cylinder.mat | ns_pretrained.pdparams | l2 error: 0.0288 |
| 8 | schrodinger | NLS.mat | NLS.mat | schrodinger_pretrained.pdparams | l2 error: 0.0735 |
注:根据 参考文献, 序号 3 的效果较差。
1. 背景简介¶
求解偏微分方程(PDE) 是一类基础的物理问题,在过去几十年里,以有限差分(FDM)、有限体积(FVM)、有限元(FEM)为代表的多种偏微分方程组数值解法趋于成熟。随着人工智能技术的高速发展,利用深度学习求解偏微分方程成为新的研究趋势。PINNs(Physics-informed neural networks) 是一种加入物理约束的深度学习网络,因此与纯数据驱动的神经网络学习相比,PINNs 可以用更少的数据样本学习到更具泛化能力的模型,其应用范围包括但不限于流体力学、热传导、电磁场、量子力学等领域。
传统的 PINNs 会将 PDE 作为 loss 的一项参与到网络训练中去,这就要求 PDE 公式为已知的先验条件,当 PDE 公式未知时,这种方法就不能实现。
DeepHPMs 着眼于 PDE 公式未知的情况,通过深度学习网络,从实验产生的高维数据中发现物理规律,即非线性 PDE 方程,并用一个深度学习网络来表征这个 PDE 方程,再将这个 PDE 网络替代传统 PINNs 方法中的 PDE 公式,对新的数据进行预测。
本问题对 Burgers, Korteweg- de Vries (KdV), Kuramoto-Sivashinsky, nonlinear Schro ̈dinger 和 Navier- Stokes equations 多种 PDE 方程进行了研究,本文档主要针对 Burgers 方程进行说明。
2. 问题定义¶
伯格斯方程(Burgers equation) 是一个模拟冲击波的传播和反射的非线性偏微分方程,该方程认为输出的解 \(u\) 与输入的位置、时间参数 \((x, t)\) 之间的关系为:
其中 \(u_t\) 为 \(u\) 对 \(t\) 的偏导数,\(u_x\) 为 \(u\) 对 \(x\) 的偏导数,\(u_{xx}\) 为 \(u\) 对 \(x\) 的二阶偏导数。
通过深度学习网络表示 PDE,即 \(u_t\) 是输入为 \(u, u_x, u_{xx}\) 的网络的输出:
3. 问题求解¶
接下来开始讲解如何将问题一步一步地转化为 PaddleScience 代码,用深度学习的方法求解该问题。为了快速理解 PaddleScience,接下来仅对模型构建、方程构建、计算域构建等关键步骤进行阐述,而其余细节请参考 API文档。
3.1 数据集介绍¶
数据集为处理好的 burgers 数据集,包含不同初始化条件下模拟数据的 \(x, t, u\) 以字典的形式存储在 .mat 文件中。
运行本问题代码前请下载 模拟数据集1 和 模拟数据集2, 下载后分别存放在路径:
3.2 模型构建¶
本问题共包含 3 个深度学习网络,分别为数据驱动的 Net1,表征 PDE 方程的 Net2 以及用于推理新数据的 Net3。
Net1 通过数据驱动的方式,使用输入的某种模拟情况 1 下的少量随机数据进行训练,学习数据规律,从而得到该模拟情况下其他所有数据的数值 \(u\)。输入为模拟情况 1 数据的 \(x, t\),输出为 \(u\),是一个\((x, t)\) 到 \(u\) 的映射函数 \(f_1: \mathbb{R}^2 \to \mathbb{R}^1\)。
对 Net1 前向推理得到的 \(u\) 值,计算其对 \(x, t\) 的偏导数 \(u_t, u_x, u_{xx}\),并将计算得到的值当作真实物理值,作为输入和标签传到 Net2 中,通过优化 loss,训练 Net2。对于 Net2,输入为 Net1 推理得到的 \(u\) 以及它对x的偏导数 \(u_x, u_{xx}\),输出为 PDE 的运算结果 \(f_{pde}\),这个值应该与 \(u_t\) 接近,即 \(u_t\) 是 \(f_{pde}\) 的 label。映射函数为 \(f_2: \mathbb{R}^3 \to \mathbb{R}^1\)。
最后,将训练好的 Net2 当作 PDE 公式,将新的模拟情况 2 下的少量数据作为输入,与 Net3 一起进行类似 PINNs 的训练,最终得到可以对模拟情况 2 进行预测的深度学习网络 Net3。对于 Net3,输入为模拟情况 2 数据的 \(x, t\),输出为 \(u\),是一个\((x, t)\) 到 \(u\) 的映射函数 \(f_3: \mathbb{R}^2 \to \mathbb{R}^1\)。
因为训练中后一个阶段网络需要使用前一个阶段网络的前向推理值,因此本问题使用 Model List 来实现,上式中 \(f_1,f_2,f_3\) 分别为一个 MLP 模型,三者共同构成了一个 Model List,用 PaddleScience 代码表示如下
注意到部分网络的输入由之前的网络计算得到,而不仅仅是数据中 \((x, t)\) 这两个变量,这也就意味着我们需要对部分网络输入进行 transform。
3.3 transform构建¶
对于 Net1,输入为 \((x, t)\) 本来不需要 transform,但由于训练中根据数据的定义域对输入数据进行了数值变换,因此同样需要transform,同样,Net3 也需要对输入进行数值变换的 transform
对于 Net2,因为它的输入为 \(u, u_x, u_{xx}\) 而 \(u\) 为其他两个网络的输出,只要进行 Net2 的前向推理,就需要 transform,因此需要两种 transform。同时,在训练 Net3 之前,需要重新注册 transform
然后依次注册 transform 后,将3 个 MLP 模型组成 Model List
注意 Net3 开始训练前,重新注册 Net2 的transform
这样我们就实例化出了一个拥有 3 个 MLP 模型,每个 MLP 包含 4 层隐藏神经元,每层神经元数为 50,使用 "sin" 作为激活函数,并包含输入 transform 的神经网络模型 model list。
3.4 参数和超参数设定¶
我们需要指定问题相关的参数,如数据集路径、输出文件路径、定义域的值等
同时需要指定训练轮数和学习率等超参数
3.5 优化器构建¶
本问题提供了两种优化器,分别为 Adam 优化器和 LBFGS 优化器,训练时只选择其中一种,需要将另一种优化器注释掉。
3.6 约束构建¶
本问题分为三个训练阶段,部分采用监督学习的方式,对 \(u\) 进行约束,部分采用无监督学习的方式,约束结果满足 PDE 公式。
无监督仍然可以采用监督约束 SupervisedConstraint,在定义约束之前,需要给监督约束指定文件路径等数据读取配置,因为数据集中没有标签数据,因此在数据读取时我们需要使用训练数据充当标签数据,并注意在之后不要使用这部分“假的”标签数据,例如
du_t 值读取了 t 的值,是“假的”标签数据。
3.6.1 第一阶段约束构建¶
第一阶段对 Net1 的训练是纯监督学习,此处采用监督约束 SupervisedConstraint
SupervisedConstraint 的第一个参数是监督约束的读取配置,配置中 “dataset” 字段表示使用的训练数据集信息,其各个字段分别表示:
name: 数据集类型,此处"IterableMatDataset"表示不分 batch 顺序读取的.mat类型的数据集;file_path: 数据集文件路径;input_keys: 输入变量名;label_keys: 标签变量名;alias_dict: 变量别名。
第二个参数是损失函数,由于是纯数据驱动,此处使用 MSE。
第三个参数是方程表达式,用于描述如何计算约束目标,计算后的值将会按照指定名称存入输出列表中,从而保证 loss 计算时可以使用这些值。
第四个参数是约束条件的名字,我们需要给每一个约束条件命名,方便后续对其索引。
在约束构建完毕之后,以我们刚才的命名为关键字,封装到一个字典中,方便后续访问。
3.6.2 第二阶段约束构建¶
第二阶段对 Net2 的训练是无监督学习,但仍可采用监督约束 SupervisedConstraint,要注意上述提到的给定“假的”标签数据
各个参数含义与 第一阶段约束构建 一致,唯一的区别是这个约束中的第二个参数,损失函数,采用 PaddleScience 预留的自定义 loss 函数类 FunctionalLoss,该类支持编写代码时自定义 loss 的计算方法,而不是使用诸如 MSE 等现有方法。本约束中的自定义 loss 函数代码请参考 自定义 loss 和 metric。
在约束构建完毕之后,以我们刚才的命名为关键字,封装到一个字典中,方便后续访问。
3.6.3 第三阶段约束构建¶
第三阶段 Net3 的训练复杂,包含了对部分初始点的监督学习、与 PDE 有关的无监督学习以及与边界条件有关的无监督学习,这里仍采用监督约束 SupervisedConstraint,同样要注意给定“假的”标签数据,各参数含义同上
在约束构建完毕之后,以我们刚才的命名为关键字,封装到一个字典中,方便后续访问。
3.7 评估器构建¶
与约束同理,虽然本问题部分采用监督学习的方式,部分采用无监督学习的方式,但仍可以使用 ppsci.validate.SupervisedValidator 构建评估器,参数含义也与约束构建,唯一的区别是评价指标 metric
3.7.1 第一阶段评估器构建¶
评价指标 metric 为 L2 正则化函数
3.7.2 第二阶段评估器构建¶
评价指标 metric 为 FunctionalMetric,这是 PaddleScience 预留的自定义 metric 函数类,该类支持编写代码时自定义 metric 的计算方法,而不是使用诸如 MSE、 L2 等现有方法。自定义 metric 函数代码请参考下一部分 自定义 loss 和 metric。
3.7.3 第三阶段评估器构建¶
因为第三阶段评价时只需要对训练得到的点的值进行评价,而不需要对边界条件满足程度或 PDE 满足程度进行评价,因此评价指标 metric 为 L2 正则化函数
3.8 自定义 loss 和 metric¶
由于本问题包含无监督学习,数据中不存在标签数据,loss 和 metric 根据 PDE 计算得到,因此需要自定义 loss 和 metric。方法为先定义相关函数,再将函数名作为参数传给 FunctionalLoss 和 FunctionalMetric。
需要注意自定义 loss 和 metric 函数的输入输出参数需要与 PaddleScience 中如 MSE 等其他函数保持一致,即输入为模型输出 output_dict 等字典变量,loss 函数输出为 loss 值 paddle.Tensor,metric 函数输出为字典 Dict[str, paddle.Tensor]。
与 PDE 相关的自定义 loss 函数为
与 PDE 相关的自定义 metric 函数为
与边界条件相关的自定义 loss 函数为
3.9 模型训练、评估¶
完成上述设置之后,只需要将上述实例化的对象按顺序传递给各个阶段 ppsci.solver.Solver,然后启动训练、评估。
第一阶段训练、评估
第二阶段训练、评估
第三阶段训练、评估
3.10 可视化¶
本问题训练结束后,可以在 evalution 中使用第三阶段网络 Net3 对模拟情况 2 的数据进行推理,结果为 \(u|_{(x,t)}\) 值,同时输出 l2 error 的值。画图部分在 plotting.py 文件中。
4. 完整代码¶
| burgers.py | |
|---|---|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136 137 138 139 140 141 142 143 144 145 146 147 148 149 150 151 152 153 154 155 156 157 158 159 160 161 162 163 164 165 166 167 168 169 170 171 172 173 174 175 176 177 178 179 180 181 182 183 184 185 186 187 188 189 190 191 192 193 194 195 196 197 198 199 200 201 202 203 204 205 206 207 208 209 210 211 212 213 214 215 216 217 218 219 220 221 222 223 224 225 226 227 228 229 230 231 232 233 234 235 236 237 238 239 240 241 242 243 244 245 246 247 248 249 250 251 252 253 254 255 256 257 258 259 260 261 262 263 264 265 266 267 268 269 270 271 272 273 274 275 276 277 278 279 280 281 282 283 284 285 286 287 288 289 290 291 292 293 294 295 296 297 298 299 300 301 302 303 304 305 306 307 308 309 310 311 312 313 314 315 316 317 318 319 320 321 322 323 324 325 326 327 328 329 330 331 332 333 334 335 336 337 338 339 340 341 342 343 344 345 346 347 348 349 350 351 352 353 354 355 356 357 358 359 360 361 362 363 364 365 366 367 368 369 370 371 372 373 374 375 376 377 378 379 380 381 382 383 384 385 386 387 388 389 390 391 392 393 394 395 396 397 398 399 400 401 402 403 404 405 406 407 408 409 410 411 412 413 414 415 416 417 418 419 420 421 422 423 424 425 426 427 428 429 430 431 432 433 434 435 436 437 438 439 440 441 | |
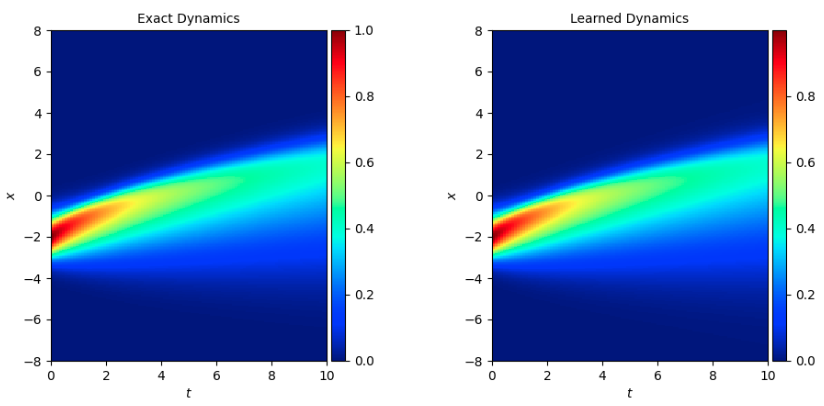
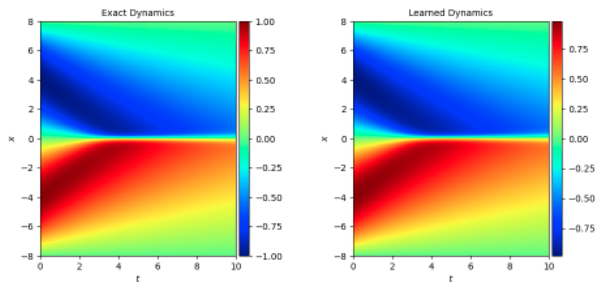
5. 结果展示¶
参考 问题定义,下图横、纵坐标分别为时间、位置参数,颜色表示 burgers 的解 u,大小参照图片右侧的颜色卡。将 burgers 方程应用在不同的问题上,u 存在不同的含义,这里可以简单的认为 u 值代表速度。
下图展示了在一定初始条件(t=0 时刻 x 对应的 u 值)下,随 t 增长,u 随 x 变化 的情况。u 的真实值和模型预测结果如下,与传统的光谱方法相比基本一致。
模拟数据集 1 是初始条件为 sin 方程时的数据集 burgers_sine.mat,模拟数据集 2 是初始条件为 exp 方程时的数据集 burgers.mat:
模拟数据集 1、2 都是初始条件为 sin 方程时的数据集 burgers_sine.mat 时:
6. 参考文献¶
创建日期: November 6, 2023