快速开始¶
本文通过一个简单的 demo 及其扩展问题,介绍如何使用 PaddleScience 训练模型,解决一类方程学习与预测问题,并可视化预测结果。
1. 问题简介¶
假设我们希望用神经网络模型去拟合 \(x \in [-\pi, \pi]\) 区间内,\(u=\sin(x)\) 这一函数。在拟合函数已知和未知两种情形下,如何去尽可能地准确拟合 \(u=\sin(x)\)。
第一种场景下,假设已知目标函数 \(u\) 的解析解就是 \(u=\sin(x)\),我们采用监督训练的思路,直接用该公式生成标签因变量 \(u\),与自变量 \(x\) 共同作为监督数据对模型进行训练。
第二种场景下,假设不知道目标函数 \(u\) 的解析解,但我们知道其满足某种微分关系,我们这里以其中一个满足条件的微分方程 \(\dfrac{\partial u} {\partial x}=\cos(x)\) 为例,介绍如何生成数据进行训练。
2. 场景一¶
目标拟合函数:
我们生成 \(N\) 组数据对 \((x_i, u_i), i=1,...,N\) 作为监督数据进行训练即可。
在撰写代码之前,我们首先导入必要的包。
然后创建日志和模型保存目录供训练过程记录和保存使用,这一步是绝大部分案例在正式开始前都需要进行的操作。
# set random seed for reproducibility
ppsci.utils.misc.set_random_seed(42)
# set output directory
OUTPUT_DIR = "./output_quick_start_case1"
# initialize logger
logger.init_logger("ppsci", f"{OUTPUT_DIR}/train.log", "info")
接下来正式开始撰写代码。
首先定义问题区间,我们使用 ppsci.geometry.Interval 定义一个线段几何形状,方便后续在该线段上对 \(x\) 进行采样。
# set input 1D-geometry([-π, π])
l_limit, r_limit = -np.pi, np.pi
x_domain = ppsci.geometry.Interval(l_limit, r_limit)
geom = {"domain": x_domain}
然后定义一个简单的 3 层 MLP 模型。
上述代码表示模型接受自变量 \(x\) 作为输入,输出预测结果 \(\hat{u}\)
然后我们定义已知的 \(u=\sin(x)\) 计算函数,作为 ppsci.constraint.InteriorConstraint 的参数,用于生成标签数据,InteriorConstraint 表示以给定的几何形状或数据集中的数据作为输入,联合给定的标签数据,指导模型进行优化。
# standard solution of sin(x)
def sin_compute_func(data: dict):
return np.sin(data["x"])
# set constraint on 1D-geometry([-π, π])
ITERS_PER_EPOCH = 100
interior_constraint = ppsci.constraint.InteriorConstraint(
output_expr={"u": lambda out: out["u"]},
label_dict={"u": sin_compute_func},
geom=geom["domain"],
dataloader_cfg={
"dataset": "NamedArrayDataset",
"iters_per_epoch": ITERS_PER_EPOCH,
"sampler": {
"name": "BatchSampler",
"shuffle": True,
},
"batch_size": 32,
},
loss=ppsci.loss.MSELoss(),
)
# wrap constraint(s) into one dict
constraint = {interior_constraint.name: interior_constraint}
此处的 interior_constraint 表示一个训练目标,即我们希望在 \([-\pi, \pi]\) 这段区间内,优化模型让模型的预测结果 \(\hat{u}\) 尽可能地接近它的标签值 \(u\)。
接下来就可以开始定义模型训练相关的内容,比如训练轮数、优化器
# set training hyper-parameters
EPOCHS = 10
# set optimizer
optimizer = ppsci.optimizer.Adam(1e-3)(model)
当训练完成,我们希望在 \([-\pi, \pi]\) 上取 1000 个点进行预测并将结果可视化,以此查看训练完毕的模型是否具备一定的预测能力。
# set visualizer
visualize_input_dict = {"x": paddle.linspace(l_limit, r_limit, 1000).reshape([1000, 1])}
visualize_input_dict["u_ref"] = paddle.sin(visualize_input_dict["x"])
visualizer = {
"visualize_u": ppsci.visualize.VisualizerScatter1D(
visualize_input_dict,
("x",),
{"u_pred": lambda out: out["u"], "u_ref": lambda out: out["u_ref"]},
prefix="u=sin(x)",
),
}
最后将上述定义的对象传递给训练调度类 Solver,即可开始模型训练
# initialize solver
solver = ppsci.solver.Solver(
model,
constraint,
OUTPUT_DIR,
optimizer,
epochs=EPOCHS,
iters_per_epoch=ITERS_PER_EPOCH,
geom=geom,
visualizer=visualizer,
)
# train model
solver.train()
训练完毕后再用刚才取的 1000 个点进行可视化
训练记录下所示
...
...
ppsci INFO: [Train][Epoch 10/10][Iter: 60/100] lr: 0.00100000, loss: 0.00084, EQ: 0.00084, batch_cost: 0.00193s, reader_cost: 0.00017s, ips: 16607.08697 samples/s, eta: 0:00:00
ppsci INFO: [Train][Epoch 10/10][Iter: 70/100] lr: 0.00100000, loss: 0.00082, EQ: 0.00082, batch_cost: 0.00193s, reader_cost: 0.00016s, ips: 16603.29541 samples/s, eta: 0:00:00
ppsci INFO: [Train][Epoch 10/10][Iter: 80/100] lr: 0.00100000, loss: 0.00078, EQ: 0.00078, batch_cost: 0.00193s, reader_cost: 0.00016s, ips: 16612.34228 samples/s, eta: 0:00:00
ppsci INFO: [Train][Epoch 10/10][Iter: 90/100] lr: 0.00100000, loss: 0.00076, EQ: 0.00076, batch_cost: 0.00193s, reader_cost: 0.00015s, ips: 16616.61847 samples/s, eta: 0:00:00
ppsci INFO: [Train][Epoch 10/10][Iter: 100/100] lr: 0.00100000, loss: 0.00075, EQ: 0.00075, batch_cost: 0.00191s, reader_cost: 0.00015s, ips: 16715.53436 samples/s, eta: 0:00:00
ppsci INFO: [Train][Epoch 10/10][Avg] loss: 0.00075, EQ: 0.00075
ppsci INFO: Finish saving checkpoint to ./output_quick_start_case1/checkpoints/latest
ppsci INFO: 1D result is saved to ./output_quick_start_case1/visual/epoch_0/u=sin(x).png
ppsci INFO: [Visualize][Epoch 0] Finished visualization.
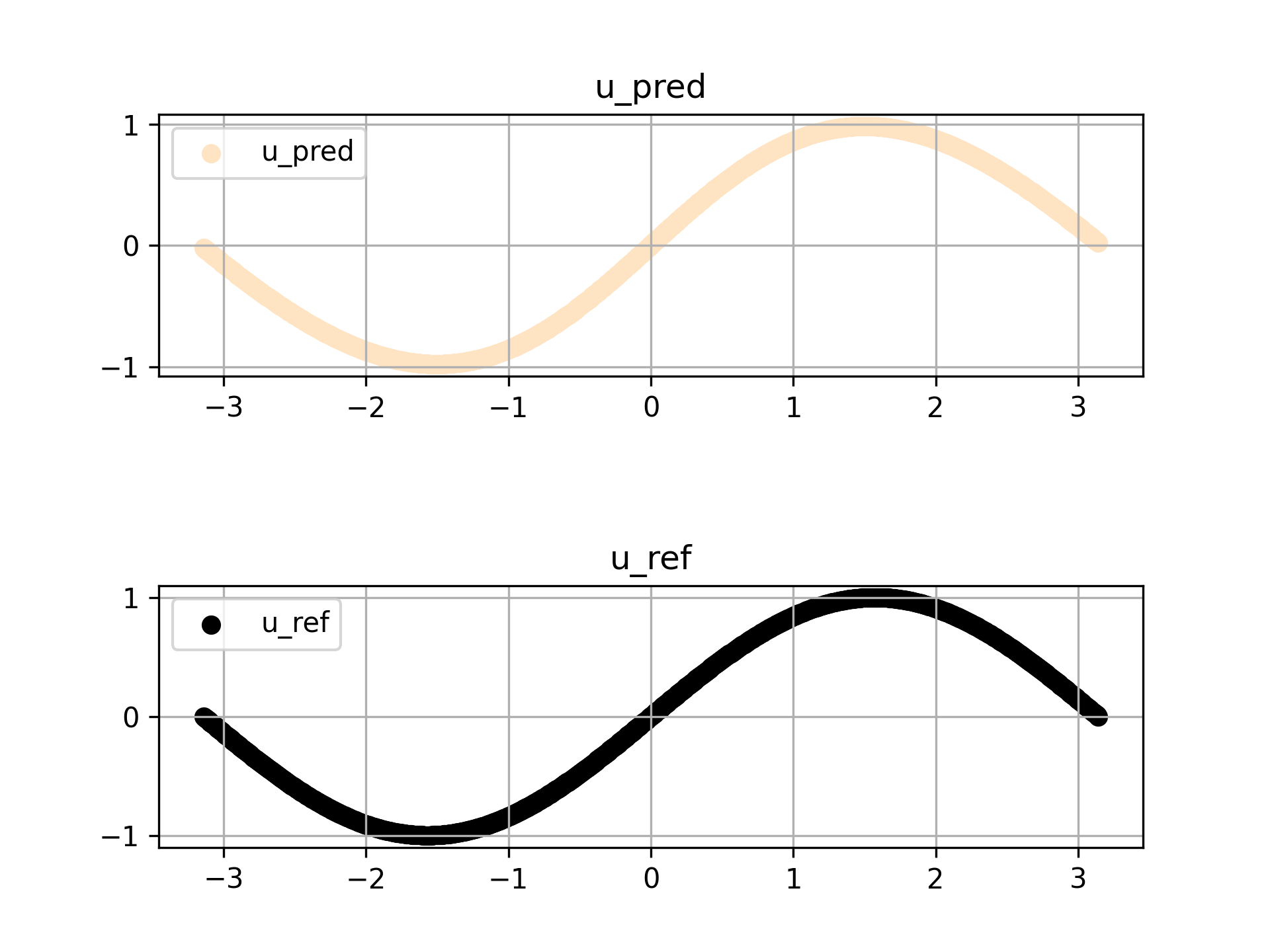
预测结果如下所示
场景一的完整代码如下所示
3. 场景二¶
可以看到场景一的监督训练方式能较好地解决函数拟合问题,但一般情况下我们是无法得知拟合函数本身的解析式的,因此也无法直接构造因变量的监督数据。
虽然无法求出解析式直接构造监督数据,但往往可以利用相关数学知识,推导出目标拟合函数符合的某种数学关系,以训练模型以满足这种数学关系的方式,达到以“间接监督”的方式优化模型的目的。
假设我们不再使用 \(u=\sin(x)\) 这一先验公式,因而无法生成标签数据 \(u\)。因此我们使用如下方程组,其含有一个偏微分方程和边界条件
构造数据对 \((x_i, \cos(x_i)), i=1,...,N\)。 这意味着我们仍然能保持模型的输入、输出不变,但优化目标变成了:让 \(\dfrac{\partial \hat{u}} {\partial x}\) 尽可能地接近 \(\cos(x)\),且 \(\hat{u}(-\pi)\) 也要尽可能地接近 \(2\)。
基于以上理论,我们对场景一的代码进行少量的改写即可得到本场景二的代码。
首先由于我们需要使用一阶微分这一操作,因此在代码开头处需导入一阶微分 API
import numpy as np
import paddle
import ppsci
from ppsci.autodiff import jacobian
from ppsci.utils import logger
然后将原来的标签生成函数改为微分关系标签生成函数
接着将 interior_constraint 这一约束条件从约束“模型输出”,改为约束“模型输出对输入的一阶微分”
interior_constraint = ppsci.constraint.InteriorConstraint(
output_expr={"du_dx": lambda out: jacobian(out["u"], out["x"])},
label_dict={"du_dx": cos_compute_func},
geom=geom["domain"],
dataloader_cfg={
"dataset": "NamedArrayDataset",
"iters_per_epoch": ITERS_PER_EPOCH,
"sampler": {
"name": "BatchSampler",
"shuffle": True,
},
"batch_size": 32,
},
loss=ppsci.loss.MSELoss(),
)
考虑到一般情况下偏微分方程的解会存在待定系数,需通过定解条件(初(边)值条件)来确定,因此需要在 interior_constraint 构建代码的后面,额外添加一个边界条件约束,如下所示
bc_constraint = ppsci.constraint.BoundaryConstraint(
{"u": lambda d: d["u"]},
{"u": lambda d: sin_compute_func(d) + 2}, # (1)
geom["domain"],
dataloader_cfg={
"dataset": "NamedArrayDataset",
"iters_per_epoch": ITERS_PER_EPOCH,
"sampler": {
"name": "BatchSampler",
"shuffle": True,
},
"batch_size": 1,
},
loss=ppsci.loss.MSELoss(),
criteria=lambda x: np.isclose(x, l_limit), # only choose x close to l_limit, i.e. -pi
)
- 对应边界条件 \(u(x_0)=sin(x_0)+2\)
然后将该边界约束添加到 constraint 中
# wrap constraint(s) into one dict
constraint = {
interior_constraint.name: interior_constraint,
bc_constraint.name: bc_constraint,
}
同样地,修改 Visualizer 绘制的标准解为 \(sin(x)+2\)
# set visualizer
visualize_input_dict = {"x": paddle.linspace(l_limit, r_limit, 1000).reshape([1000, 1])}
visualize_input_dict["u_ref"] = paddle.sin(visualize_input_dict["x"]) + 2
visualizer = {
"visualize_u": ppsci.visualize.VisualizerScatter1D(
visualize_input_dict,
("x",),
{"u_pred": lambda out: out["u"], "u_ref": lambda out: out["u_ref"]},
prefix="u=sin(x)+2",
),
}
修改完毕后执行训练
# initialize solver
solver = ppsci.solver.Solver(
model,
constraint,
OUTPUT_DIR,
optimizer,
epochs=EPOCHS,
iters_per_epoch=ITERS_PER_EPOCH,
geom=geom,
visualizer=visualizer,
)
# train model
solver.train()
训练日志如下所示
...
...
ppsci INFO: [Train][Epoch 10/10][Iter: 70/100] lr: 0.00100000, loss: 0.00035, EQ: 0.00035, batch_cost: 0.01183s, reader_cost: 0.00017s, ips: 2705.18917 samples/s, eta: 0:00:00
ppsci INFO: [Train][Epoch 10/10][Iter: 80/100] lr: 0.00100000, loss: 0.00035, EQ: 0.00035, batch_cost: 0.01133s, reader_cost: 0.00017s, ips: 2823.74760 samples/s, eta: 0:00:00
ppsci INFO: [Train][Epoch 10/10][Iter: 90/100] lr: 0.00100000, loss: 0.00036, EQ: 0.00036, batch_cost: 0.01141s, reader_cost: 0.00017s, ips: 2803.77351 samples/s, eta: 0:00:00
ppsci INFO: [Train][Epoch 10/10][Iter: 100/100] lr: 0.00100000, loss: 0.00036, EQ: 0.00036, batch_cost: 0.01106s, reader_cost: 0.00016s, ips: 2892.93859 samples/s, eta: 0:00:00
ppsci INFO: [Train][Epoch 10/10][Avg] loss: 0.00036, EQ: 0.00036
ppsci INFO: Finish saving checkpoint to ./output_quick_start_case2/checkpoints/latest
ppsci INFO: 1D result is saved to ./output_quick_start_case2/visual/epoch_0/u=sin(x)+2.png
ppsci INFO: [Visualize][Epoch 0] Finished visualization.
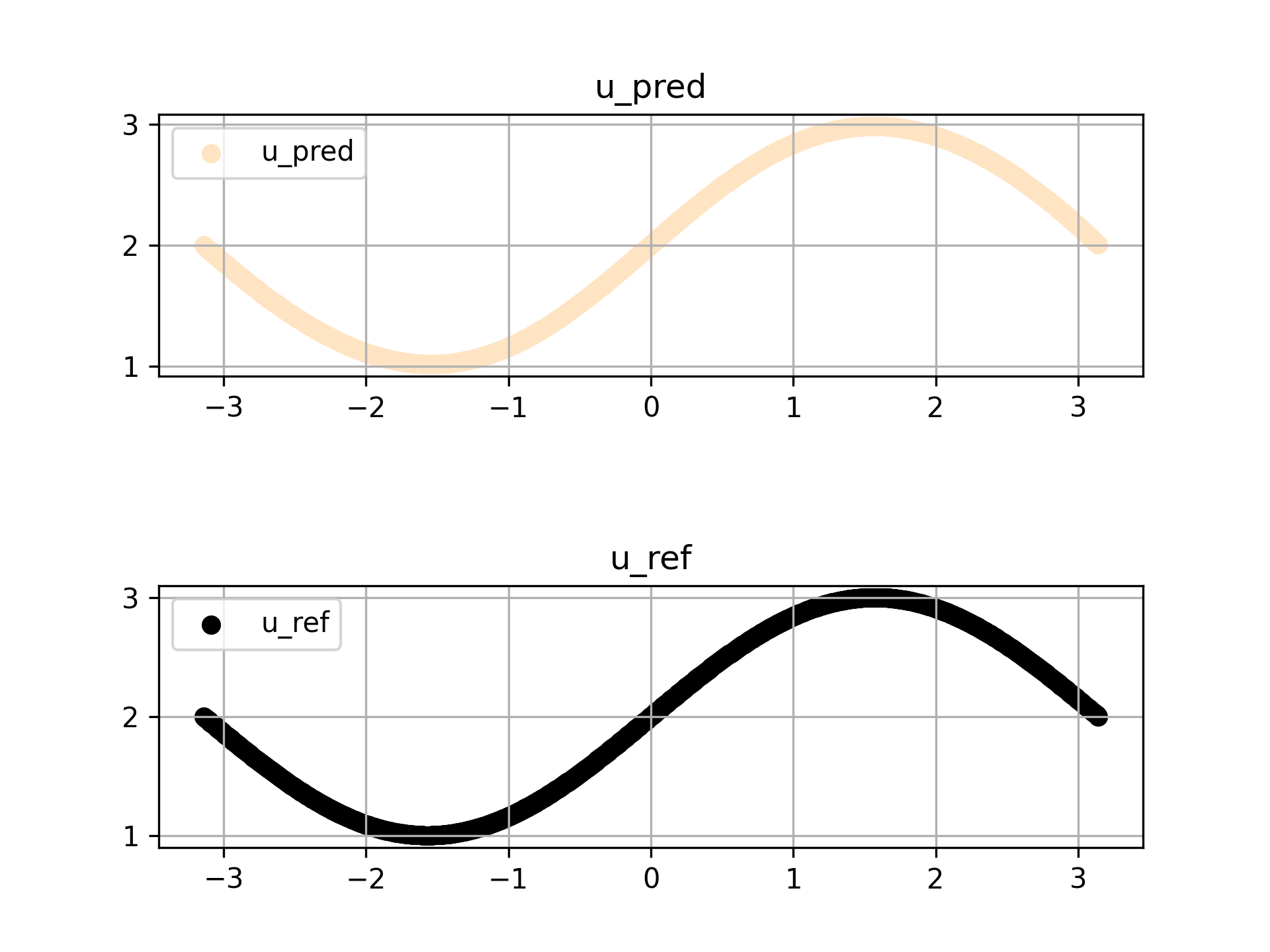
预测结果如下所示
可以发现利用微分关系训练的模型仍然具备良好的预测能力,并且结合定解条件,能学习出同时符合微分方程和定解条件的正确解模型。
场景二的完整代码如下所示
| quickstart_case2.py | |
|---|---|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 | |
创建日期: November 6, 2023