Chip Heat Simulation¶
| 预训练模型 | 指标 |
|---|---|
| chip_heat_pretrained.pdparams | MSE.chip(down_mse): 0.04177 MSE.chip(left_mse): 0.01783 MSE.chip(right_mse): 0.03767 MSE.chip(top_mse): 0.05034 |
1. 背景简介¶
芯片热仿真研究主要聚焦于预测和分析集成电路(IC)在操作过程中的温度分布,以及热效应对芯片性能、功耗、可靠性和寿命的影响。随着电子设备向更高性能、更高密度和更小尺寸发展,热管理成为芯片设计和制造中的一个关键挑战。
芯片热仿真研究为理解和解决芯片热管理问题提供了重要工具和方法,对于提高芯片的性能、降低功耗、保证可靠性和延长寿命有着至关重要的作用。随着电子设备朝着更高性能和更紧凑的方向发展,热仿真研究的重要性将会进一步增加。
芯片热仿真在工程和科学领域具有多方面的重要性,主要体现在以下几个方面:
- 设计优化和验证: 芯片热仿真可以帮助工程师和科学家在设计初期评估不同结构和材料的热特性,以优化设计并验证其可靠性。通过仿真模拟不同工作负载下的温度分布和热传导效应,可以提前发现潜在的热问题并进行针对性的改进,从而降低后期开发成本和风险。
- 热管理和散热设计: 芯片热仿真可以帮助设计有效的热管理系统和散热方案,以确保芯片在长时间高负载运行时保持在安全的工作温度范围内。通过分析芯片周围的散热结构、风扇配置、散热片设计等因素,可以优化热传导和散热效率,提高系统的稳定性和可靠性。
- 性能预测和优化: 温度对芯片的性能和稳定性有重要影响。芯片热仿真可以帮助预测芯片在不同工作负载和环境条件下的性能表现,包括处理器速度、功耗和电子器件的寿命等方面。通过对热效应的建模和分析,可以优化芯片的设计和工作条件,以实现更好的性能和可靠性。
- 节能和环保: 有效的热管理和散热设计可以降低系统能耗,提高能源利用效率,从而实现节能和环保的目标。通过减少系统中热量的损失和浪费,可以降低能源消耗和碳排放,减少对环境的负面影响。
综上所述,芯片热仿真在工程和科学领域中具有重要的作用和价值,可以帮助优化设计、提高性能、降低成本、保护环境等方面取得积极的效果。
2. 问题定义¶
2.1 问题描述¶
为了搭建通用的热仿真模型,我们首先对一般情况下热仿真问题进行简要描述,热仿真旨在通过全局求解热传导方程来预测给定物体的温度场,通常可以通过以下控制方程来进行表示:
其中 \(\Omega\subset \mathbb{R}^{n},~n=1,2,3\) 为给定物体材料的模拟区域,如图所示为一个具有随机热源分布的2D芯片模拟区域。\(T(x,t),~S(x,t)\) 分别表示在任意时空位置 \((x,t)\) 处温度和热源分布,\(t_*\) 为温度阈值。这里 \(k\)、\(\rho\)、\(c_p\) 均为给定物体的材料特性,分别表示材料传热系数、质量密度和比热容。为了方便,我们关注给定物体材料的静态温度场,并通过设置 \(\frac{dT}{dt}=0\) 来简化方程:
对于给定物体材料的通用热仿真模型,除了要满足控制方程(1),其温度场还取决于一些关键的 PDE 配置,包括但不限于材料特性和几何参数等。
第一类 PDE 配置是给定物体材料的边界条件:
- Dirichlet边界条件: 表面上的温度场固定为 \(q_d\):
- Neumann边界条件: 表面上的温度通量是固定为 \(q_n\),当 \(q_n =0\) 时,表明表面完全绝缘,称为绝热边界条件。
- 对流边界条件:也称为牛顿边界条件,该边界条件对应于表面相同方向上的热传导和对流之间的平衡,其中 \(h\) 和 \(T_{amb}\) 代表表面的对流系数和环境温度。
- 辐射边界条件:该边界条件对应于表面上由温差产生的电磁波辐射,其中 \(\epsilon\) 和 \(\sigma\) 分别代表热辐射系数和Stefan-Boltzmann系数。
第二类PDE配置是给定物体材料的边界或内部热源的位置和强度。本工作考虑了以下两种类型的热源:
- 边界随机热源:由 Neumann 边界条件(2)定义,此时 \(q_n\) 为关于 \(x\) 的函数,即任意给定的温度通量分布;
- 内部随机热源:由控制方程(1)定义,此时 \(S(x)\) 为关于 \(x\) 的函数,即任意给定的热源分布。
我们的目的是,在给定的物体材料的通用热仿真模型上,输入任意的第一类或第二类设计配置,我们均可以得到对应的温度场分布情况,在边界上我们任意指定边界类型和参数。值得注意的是,这项工作中开发的通用热仿真的 PI-DeepONet 方法并不限于 第一类或第二类设计配置 条件和规则的几何形状。通过超出当前工作范围的进一步代码修改,它们可以应用于各种载荷、材料属性,甚至各种不规则的几何形状。
2.2 PI-DeepONet模型¶
PI-DeepONet模型,将 DeepONet 和 PINN 方法相结合,是一种结合了物理信息和算子学习的深度神经网络模型。这种模型可以通过控制方程的物理信息来增强 DeepONet 模型,同时可以将不同的 PDE 配置分别作为不同的分支网络的输入数据,从而可以有效地用于在各种(参数和非参数)PDE 配置下进行超快速的模型预测。
对于芯片热仿真问题,PI-DeepONet 模型可以表示为如图所示的模型结构:
如图所示,我们一共使用了 3 个分支网络和一个主干网络,分支网络分别输入边界类型指标、随机热源分布 \(S(x, y)\) 和边界函数 \(Q(x, y)\),主干网络输入二维坐标点坐标信息。每个分支网和主干网均输出 \(q\) 维特征向量,通过 Hadamard(逐元素)乘积组合所有这些输出特征,然后将所得向量相加为预测温度场的标量输出。
3. 问题求解¶
接下来开始讲解如何将该问题一步一步地转化为 PaddleScience 代码,用深度学习的方法求解该换热器热仿真问题。为了快速理解 PaddleScience,接下来仅对模型构建、约束构建等关键步骤进行阐述,而其余细节请参考API文档。
3.1 模型构建¶
在芯片热仿真问题中,每一个已知的坐标点 \((x, y)\) 和每一组边界类型 \(bt\)、随机热源分布 \(S(x, y)\) 以及边界函数 \(Q(x, y)\) 都对应一组芯片的温度分布 \(T\),一个待求解的未知量。我们在这里使用 3 个分支网络和一个主干网络,4 个网络均为 MLP(Multilayer Perceptron, 多层感知机) 。 3 个分支网络分别表示 \((bt, S, Q)\) 到输出函数 \((b_1, b_2, b_3)\) 的映射函数 \(f_1,f_2,f_3: \mathbb{R}^3 \to \mathbb{R}^{q}\),即:
上式中 \(f_1, f_2, f_3\) 均为 MLP 模型,\((b_1,b_2,b_3)\) 分别为三个分支网络的输出函数,\(q\) 为输出函数的维数。主干网络表示 \((x, y)\) 到输出函数 \(t_0\) 的映射函数 \(f_4: \mathbb{R} \to \mathbb{R}^{q}\),即:
上式中 \(f_4\) 为 MLP 模型,\((t_0)\) 为主支网络的输出函数,\(q\) 为输出函数的维数。我们可以将三个分支网络和主干网络的输出函数 \((b_1, b_2, b_3, t_0)\) 进行 Hadamard(逐元素)乘积再相加得到标量温度场,即:
我们定义 PaddleScience 内置的 ChipHeats 模型类,并调用,PaddleScience 代码表示如下
这样我们就实例化出了一个拥有 4 个 MLP 模型的 ChipHeats 模型,每个分支网络包含 9 层隐藏神经元,每层神经元数为 256,主干网络包含 6 层隐藏神经元,每层神经元数为 128,使用 "Swish" 作为激活函数,并包含一个输出函数 \(T\) 的神经网络模型 model。更多相关内容请参考文献 A fast general thermal simulation model based on MultiBranch Physics-Informed deep operator neural network。
3.2 计算域构建¶
对本文中芯片热仿真问题构造训练区域,即以 \([0, 1]\times[0, 1]\) 的二维区域,该区域可以直接使用 PaddleScience 内置的空间几何 Rectangle来构造计算域。代码如下
提示
Rectangle 和 TimeDomain 是两种可以单独使用的 Geometry 派生类。
如输入数据只来自于二维矩形几何域,则可以直接使用 ppsci.geometry.Rectangle(...) 创建空间几何域对象;
如输入数据只来自一维时间域,则可以直接使用 ppsci.geometry.TimeDomain(...) 构建时间域对象。
3.3 输入数据构建¶
使用二维相关且尺度不变的高斯随机场来生成随机热源分布 \(S(x)\) 和边界函数 \(Q(x)\)。我们参考 gaussian-random-fields 中描述的Python实现,其中相关性由无标度谱来解释,即
采样函数的平滑度由长度尺度系数 \(\alpha\) 决定,\(\alpha\) 值越大,得到的随机热源分布 \(S(x)\) 和边界函数 \(Q(x)\) 越平滑。在本文我们采用 \(\alpha = 4\)。还可以调整该参数以生成类似于特定优化任务中的热源分布 \(S(x)\) 和边界函数 \(Q(x)\)。
通过高斯随机场来生成随机热源分布 \(S(x)\) 和边界函数 \(Q(x)\)的训练和测试输入数据。代码如下
然后对训练数据和测试数据按照空间坐标进行分类,将训练数据和测试数据分类成左边、右边、上边、下边以及内部数据。代码如下
97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136 137 138 139 140 141 142 143 144 145 146 147 148 149 150 151 152 153 154 155 156 157 158 159 160 161 162 163 164 165 166 167 168 169 170 171 172 173 174 175 176 177 178 179 180 181 182 183 184 185 186 187 188 189 190 191 192 | |
3.4 约束构建¶
在构建约束之前,需要先介绍一下ChipHeatDataset,它继承自 Dataset 类,可以迭代的读取由不同 numpy.ndarray 组成的数组数据集。由于所用的模型分支网数目较多,所用的数据量较大。若先对数据进行组合,将导致输入数据占用的内存很大,因此采用 ChipHeatDataset 迭代读取数据。
芯片热仿真问题由 2.1 问题描述 中描述的方程组成,此时我们对左边、右边、上边、下边以及内部数据分别设置五个约束条件,接下来使用 PaddleScience 内置的 SupervisedConstraint 构建上述四种约束条件,代码如下
194 195 196 197 198 199 200 201 202 203 204 205 206 207 208 209 210 211 212 213 214 215 216 217 218 219 220 221 222 223 224 225 226 227 228 229 230 231 232 233 234 235 236 237 238 239 240 241 242 243 244 245 246 247 248 249 250 251 252 253 254 255 256 257 258 259 260 261 262 263 264 265 266 267 268 269 270 271 272 273 274 275 276 277 278 279 280 281 282 283 284 285 286 287 288 289 290 291 292 293 294 295 296 297 298 299 300 301 302 303 304 305 306 307 308 309 310 311 312 313 314 315 316 317 318 319 320 321 322 323 324 325 326 327 328 329 330 331 332 333 334 335 336 337 338 339 340 341 342 343 344 345 346 347 348 349 350 351 352 353 354 355 356 357 358 359 360 361 362 363 364 365 366 367 368 369 370 371 372 373 374 375 376 377 378 379 380 381 | |
SupervisedConstraint 的第一个参数是监督约束的读取配置,其中 “dataset” 字段表示使用的训练数据集信息,各个字段分别表示:
name: 数据集类型,此处ChipHeatDataset表示分 batch 顺序迭代的读取数据;input: 输入变量名;label: 标签变量名;index: 输入数据集的索引;data_type: 输入数据的类型;weight: 权重大小。
"sampler" 字段定义了使用的 Sampler 类名为 BatchSampler,另外还指定了该类初始化时参数 drop_last 为 False、shuffle 为 True。
第二个参数是损失函数,此处我们选用常用的 MSE 函数,且 reduction 为 "mean",即我们会将参与计算的所有数据点产生的损失项求和取平均;
第三个参数是标签表达式列表,此处我们使用与左边、右边、上边、下边以及内部区域相对应的方程表达式,同时我们分别用 \(0,1,2,3\) 代表Dirichlet边界、Neumann 边界、对流边界以及辐射边界,对与不同的边界类型,设置不同的边界条件;
第四个参数是约束条件的名字,我们需要给每一个约束条件命名,方便后续对其索引。
在微分方程约束和监督约束构建完毕之后,以我们刚才的命名为关键字,封装到一个字典中,方便后续访问。
3.5 优化器构建¶
接下来我们需要指定学习率,学习率设为 0.001,训练过程会调用优化器来更新模型参数,此处选择较为常用的 Adam 优化器。
3.6 评估器构建¶
在训练过程中通常会按一定轮数间隔,用验证集(测试集)评估当前模型的训练情况,我们使用 ppsci.validate.SupervisedValidator 构建评估器。
394 395 396 397 398 399 400 401 402 403 404 405 406 407 408 409 410 411 412 413 414 415 416 417 418 419 420 421 422 423 424 425 426 427 428 429 430 431 432 433 434 435 436 437 438 439 440 441 442 443 444 445 446 447 448 449 450 451 452 453 454 455 456 457 458 459 460 461 462 463 464 465 466 467 468 469 470 471 472 473 474 475 476 477 478 479 480 481 482 483 484 485 486 487 488 489 490 491 492 493 494 495 | |
配置与 3.4 约束构建 的设置类似。需要注意的是,由于评估所用的数据量不是很多,因此我们不需要使用ChipHeatDataset 迭代的读取数据,在这里使用NamedArrayDataset 读取数据。
3.7 模型训练¶
完成上述设置之后,只需要将上述实例化的对象按顺序传递给 ppsci.solver.Solver,然后启动训练、评估。
3.8 结果可视化¶
最后在给定的可视化区域上进行预测并可视化,可视化数据是区域内的二维点集,每个坐标 \((x, y)\) 处,对应的温度值 \(T\),在此我们画出 \(T\) 在区域上的变化图像。同时可以根据需要,设置不同的边界类型、随机热源分布 \(S(x)\) 和边界函数 \(Q(x)\),代码如下:
4. 完整代码¶
| chip_heat.py | |
|---|---|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136 137 138 139 140 141 142 143 144 145 146 147 148 149 150 151 152 153 154 155 156 157 158 159 160 161 162 163 164 165 166 167 168 169 170 171 172 173 174 175 176 177 178 179 180 181 182 183 184 185 186 187 188 189 190 191 192 193 194 195 196 197 198 199 200 201 202 203 204 205 206 207 208 209 210 211 212 213 214 215 216 217 218 219 220 221 222 223 224 225 226 227 228 229 230 231 232 233 234 235 236 237 238 239 240 241 242 243 244 245 246 247 248 249 250 251 252 253 254 255 256 257 258 259 260 261 262 263 264 265 266 267 268 269 270 271 272 273 274 275 276 277 278 279 280 281 282 283 284 285 286 287 288 289 290 291 292 293 294 295 296 297 298 299 300 301 302 303 304 305 306 307 308 309 310 311 312 313 314 315 316 317 318 319 320 321 322 323 324 325 326 327 328 329 330 331 332 333 334 335 336 337 338 339 340 341 342 343 344 345 346 347 348 349 350 351 352 353 354 355 356 357 358 359 360 361 362 363 364 365 366 367 368 369 370 371 372 373 374 375 376 377 378 379 380 381 382 383 384 385 386 387 388 389 390 391 392 393 394 395 396 397 398 399 400 401 402 403 404 405 406 407 408 409 410 411 412 413 414 415 416 417 418 419 420 421 422 423 424 425 426 427 428 429 430 431 432 433 434 435 436 437 438 439 440 441 442 443 444 445 446 447 448 449 450 451 452 453 454 455 456 457 458 459 460 461 462 463 464 465 466 467 468 469 470 471 472 473 474 475 476 477 478 479 480 481 482 483 484 485 486 487 488 489 490 491 492 493 494 495 496 497 498 499 500 501 502 503 504 505 506 507 508 509 510 511 512 513 514 515 516 517 518 519 520 521 522 523 524 525 526 527 528 529 530 531 532 533 534 535 536 537 538 539 540 541 542 543 544 545 546 547 548 549 550 551 552 553 554 555 556 557 558 559 560 561 562 563 564 565 566 567 568 569 570 571 572 573 574 575 576 577 578 579 580 581 582 583 584 585 586 587 588 589 590 591 592 593 594 595 596 597 598 599 600 601 602 603 604 605 606 607 608 609 610 611 612 613 614 615 616 617 618 619 620 621 622 623 624 625 626 627 628 629 630 631 632 633 634 635 636 637 638 639 640 641 642 643 644 645 646 647 648 649 650 651 652 653 654 655 656 657 658 659 660 661 662 663 664 665 666 667 668 669 670 671 672 673 674 675 676 677 678 679 680 681 682 683 684 685 686 687 688 689 690 691 692 693 694 695 696 697 698 699 700 701 702 703 704 705 706 707 708 709 710 711 712 713 714 715 716 717 718 719 720 721 722 723 724 725 726 727 728 729 730 731 732 733 734 735 736 737 738 739 740 741 742 743 744 745 746 747 748 749 | |
5. 结果展示¶
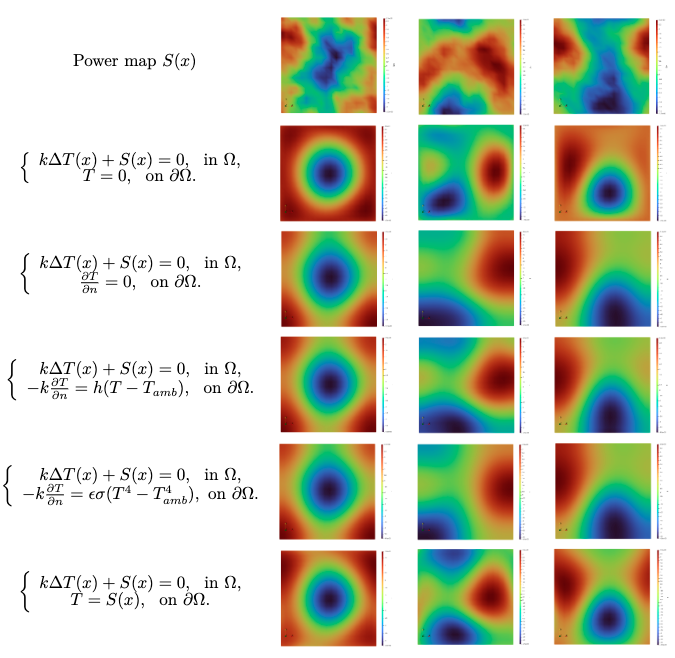
通过高斯随机场生成三组随机热源分布 \(S(x)\),如图中第一行所示。接下来我们可以设置第一类 PDE 中的任意边界条件,在此我们给出了五类边界条件,如图中第一列控制方程中边界方程所示,在测试过程中,我们设 \(k = 100,~h = 100,~T_{amb} = 1,~\epsilon\sigma= 5.6 \times 10^{-7}\)。 在不同随机热源 \(S(x)\) 分布和不同边界条件下,我们通过 PI-DeepONet 模型测试的温度场分布如图所示。从图中可知,尽管随机热源分布 \(S(x)\) 和边界条件在测试样本之间存在着显着差异,但 PI-DeepONet 模型均可以正确预测由热传导方程控制的内部和边界上的二维扩散性质解。
6. 参考资料¶
参考代码: gaussian-random-fields